!python --versionPython 3.7.15Tabular as a Form of Function Approximation
Kobus Esterhuysen
July 14, 2022
FunctionApproxWe now take a step back. Let us again consider a finite set of
We refer to this kind of setup as Tabular - each
We make use of the approach followed in http://web.stanford.edu/class/cme241/.
from __future__ import annotations
from typing import Tuple, Sequence, Iterator, List, TypeVar, Generic, overload
from scipy.stats import norm
import numpy as np
from abc import ABC, abstractmethod
from collections import defaultdict
from dataclasses import dataclass, replace, field
import itertoolsLet’s setup a Data Model:
def example_model_data_generator() -> Iterator[DataSeq]:
coeffs: Aug_Triple = (2., 10., 4., -6.)
values = np.linspace(-10.0, 10.0, 21)
pts: Sequence[Triple] = [(x, y, z) for x in values for y in values
for z in values]
d = norm(loc=0., scale=2.0) #. normal distribution
while True:
res: List[Tuple[Triple, float]] = []
for pt in pts:
# pt: np.ndarray = np.random.randn(3)
x_val: Triple = (pt[0], pt[1], pt[2])
y_val: float = coeffs[0] + np.dot(coeffs[1:], pt) + \
d.rvs(size=1)[0]
res.append((x_val, y_val))
yield res<generator object example_model_data_generator at 0x7f037d757b50>[((-10.0, -10.0, -10.0), -79.48767732128363),
((-10.0, -10.0, -9.0), -82.06969223580259),
((-10.0, -10.0, -8.0), -88.14484959324732),
((-10.0, -10.0, -7.0), -98.79564553233163),
((-10.0, -10.0, -6.0), -100.54401550033428),
((-10.0, -10.0, -5.0), -108.94733942986316),
((-10.0, -10.0, -4.0), -114.67929646511288),
((-10.0, -10.0, -3.0), -122.14685477082418),
((-10.0, -10.0, -2.0), -127.84849723837867),
((-10.0, -10.0, -1.0), -131.72740725100914)]We define the next class to implement the function approximation X to allow for arbitrary data types
X = TypeVar('X') #. for arbitrary data types scriptX
F = TypeVar('F', bound='FunctionApprox')
SMALL_NUM = 1e-6
class FunctionApprox(ABC, Generic[X]):
@abstractmethod
def __add__(self: F, other: F) -> F:
pass
@abstractmethod
def __mul__(self: F, scalar: float) -> F:
pass
@abstractmethod
def objective_gradient(
self: F,
xy_vals_seq: Iterable[Tuple[X, float]],
obj_deriv_out_fun: Callable[[Sequence[X], Sequence[float]], np.ndarray]
) -> Gradient[F]:
pass
@abstractmethod
def evaluate(self, x_values_seq: Iterable[X]) -> np.ndarray:
pass
def __call__(self, x_value: X) -> float:
return self.evaluate([x_value]).item()
@abstractmethod
def update_with_gradient(
self: F,
gradient: Gradient[F]
) -> F:
pass
def update(
self: F,
xy_vals_seq: Iterable[Tuple[X, float]]
) -> F:
def deriv_func(x: Sequence[X], y: Sequence[float]) -> np.ndarray:
return self.evaluate(x) - np.array(y)
return self.update_with_gradient(
self.objective_gradient(xy_vals_seq, deriv_func)
)
@abstractmethod
def solve(
self: F,
xy_vals_seq: Iterable[Tuple[X, float]],
error_tolerance: Optional[float] = None
) -> F:
pass
@abstractmethod
def within(self: F, other: F, tolerance: float) -> bool:
pass
def iterate_updates(
self: F,
xy_seq_stream: Iterator[Iterable[Tuple[X, float]]]
) -> Iterator[F]:
return iterate.accumulate(
xy_seq_stream,
lambda fa, xy: fa.update(xy),
initial=self
)
def rmse(
self,
xy_vals_seq: Iterable[Tuple[X, float]]
) -> float:
x_seq, y_seq = zip(*xy_vals_seq)
errors: np.ndarray = self.evaluate(x_seq) - np.array(y_seq)
return np.sqrt(np.mean(errors * errors))
def argmax(self, xs: Iterable[X]) -> X:
args: Sequence[X] = list(xs)
return args[np.argmax(self.evaluate(args))]Now we implement the main class of this project: Tabular. A tabular prediction fits well into the interface of FunctionApprox. By considering tabular prediction as a special case of the class FunctionApprox, we can cast the
Tabular class.The Tabular class approximates a function with a discrete domain (X), without interpolation. The value for each X is maintained as a weighted average of observations based on recency (tracked by count_to_weight_func).
So here is the implementation of this class:
@dataclass(frozen=True)
class Tabular(FunctionApprox[X]):
values_map: Mapping[X, float] = field(default_factory=lambda: {})
counts_map: Mapping[X, int] = field(default_factory=lambda: {})
count_to_weight_func: Callable[[int], float] = \
field(default_factory=lambda: lambda n: 1.0 / n)
def objective_gradient( #. scriptG(w_t)
self,
xy_vals_seq: Iterable[Tuple[X, float]],
obj_deriv_out_fun: Callable[[Sequence[X], Sequence[float]], float]
) -> Gradient[Tabular[X]]:
x_vals, y_vals = zip(*xy_vals_seq)
obj_deriv_out: np.ndarray = obj_deriv_out_fun(x_vals, y_vals)
sums_map: Dict[X, float] = defaultdict(float)
counts_map: Dict[X, int] = defaultdict(int)
for x, o in zip(x_vals, obj_deriv_out):
sums_map[x] += o
counts_map[x] += 1
return Gradient(replace(
self,
values_map={x: sums_map[x] / counts_map[x] for x in sums_map},
counts_map=counts_map
))
def __add__(self, other: Tabular[X]) -> Tabular[X]:
values_map: Dict[X, float] = {}
counts_map: Dict[X, int] = {}
for key in set.union(
set(self.values_map.keys()),
set(other.values_map.keys())
):
values_map[key] = self.values_map.get(key, 0.) + \
other.values_map.get(key, 0.)
counts_map[key] = counts_map.get(key, 0) + \
other.counts_map.get(key, 0)
return replace(
self,
values_map=values_map,
counts_map=counts_map
)
def __mul__(self, scalar: float) -> Tabular[X]:
return replace(
self,
values_map={x: scalar*y for x, y in self.values_map.items()}
)
def evaluate(self, x_values_seq: Iterable[X]) -> np.ndarray:
return np.array([self.values_map.get(x, 0.) for x in x_values_seq])
def update_with_gradient(
self,
gradient: Gradient[Tabular[X]]
) -> Tabular[X]:
values_map: Dict[X, float] = dict(self.values_map)
counts_map: Dict[X, int] = dict(self.counts_map)
for key in gradient.function_approx.values_map:
counts_map[key] = counts_map.get(key, 0) + \
gradient.function_approx.counts_map[key]
weight: float = self.count_to_weight_func(counts_map[key])
values_map[key] = values_map.get(key, 0.) - \
weight * gradient.function_approx.values_map[key]
return replace(
self,
values_map=values_map,
counts_map=counts_map
)
def solve(
self,
xy_vals_seq: Iterable[Tuple[X, float]],
error_tolerance: Optional[float] = None
) -> Tabular[X]:
values_map: Dict[X, float] = {}
counts_map: Dict[X, int] = {}
for x, y in xy_vals_seq:
counts_map[x] = counts_map.get(x, 0) + 1
weight: float = self.count_to_weight_func(counts_map[x])
values_map[x] = weight * y + (1 - weight) * values_map.get(x, 0.)
return replace(
self,
values_map=values_map,
counts_map=counts_map
)
def within(self, other: FunctionApprox[X], tolerance: float) -> bool:
if isinstance(other, Tabular):
return all(abs(self.values_map[s] - other.values_map.get(s, 0.))
<= tolerance for s in self.values_map)
return FalseThe attributes are: - values_map: Mapping[X, float] - mapping from each counts_map: Mapping[X, int] - mapping from each count_to_weight_func: Callable[[int], float] - defines a function from number of
The most important methods are: - solve - calculate the average of all the xy_vals_seq: Iterable[Tuple[X, float]] - error_tolerance: Optional[float]=None - output - Tabular[X] - update - Update the FunctionApprox’s internal parameters xy_vals_seq: Iterable[Tuple[X, float]]
- the F - the FunctionApprox evaluate - Evaluate the function approximation by looking up the value in the mapping for each state. If an X value has not been seen before and hence not initialized,returns 0 - inputs - x_values_seq: Iterable[X] - output - np.ndarray
The Gradient class holds the FunctionApprox F:
@dataclass(frozen=True)
class Gradient(Generic[F]):
function_approx: F
@overload
def __add__(self, x: Gradient[F]) -> Gradient[F]:
...
@overload
def __add__(self, x: F) -> F:
...
def __add__(self, x):
if isinstance(x, Gradient):
return Gradient(self.function_approx + x.function_approx)
return self.function_approx + x
def __mul__(self: Gradient[F], x: float) -> Gradient[F]:
return Gradient(self.function_approx*x)
def zero(self) -> Gradient[F]:
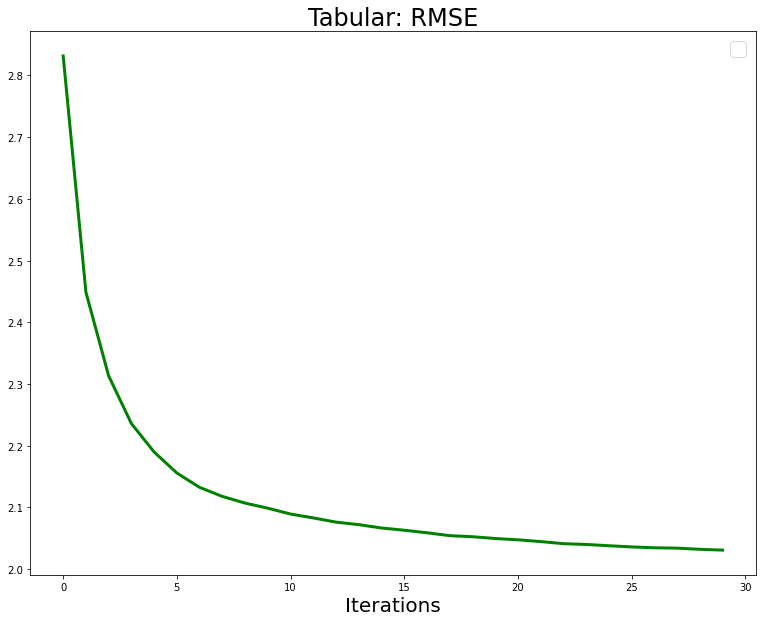
return Gradient(self.function_approx*0.0)Next we run a training loop and plot the RMSE.
tabular: Tabular[Triple] = Tabular()
rmses = []
for xy_seq in itertools.islice(data_gen, trn_iterations):
tabular = tabular.update(xy_seq)
rmse: float = tabular.rmse(tst_data)
rmses.append(rmse)
print(f"RMSE: {rmse:.2f}")RMSE: 2.83
RMSE: 2.45
RMSE: 2.31
RMSE: 2.24
RMSE: 2.19
RMSE: 2.16
RMSE: 2.13
RMSE: 2.12
RMSE: 2.11
RMSE: 2.10
RMSE: 2.09
RMSE: 2.08
RMSE: 2.08
RMSE: 2.07
RMSE: 2.07
RMSE: 2.06
RMSE: 2.06
RMSE: 2.05
RMSE: 2.05
RMSE: 2.05
RMSE: 2.05
RMSE: 2.04
RMSE: 2.04
RMSE: 2.04
RMSE: 2.04
RMSE: 2.04
RMSE: 2.03
RMSE: 2.03
RMSE: 2.03
RMSE: 2.03import matplotlib.pyplot as plt
fig,axs = plt.subplots(figsize=(13,10))
axs.set_xlabel('Iterations', fontsize=20)
axs.set_title(f'Tabular: RMSE', fontsize=24)
axs.plot(rmses, linewidth=3.0, color='green')
axs.legend(fontsize=20);WARNING:matplotlib.legend:No handles with labels found to put in legend.